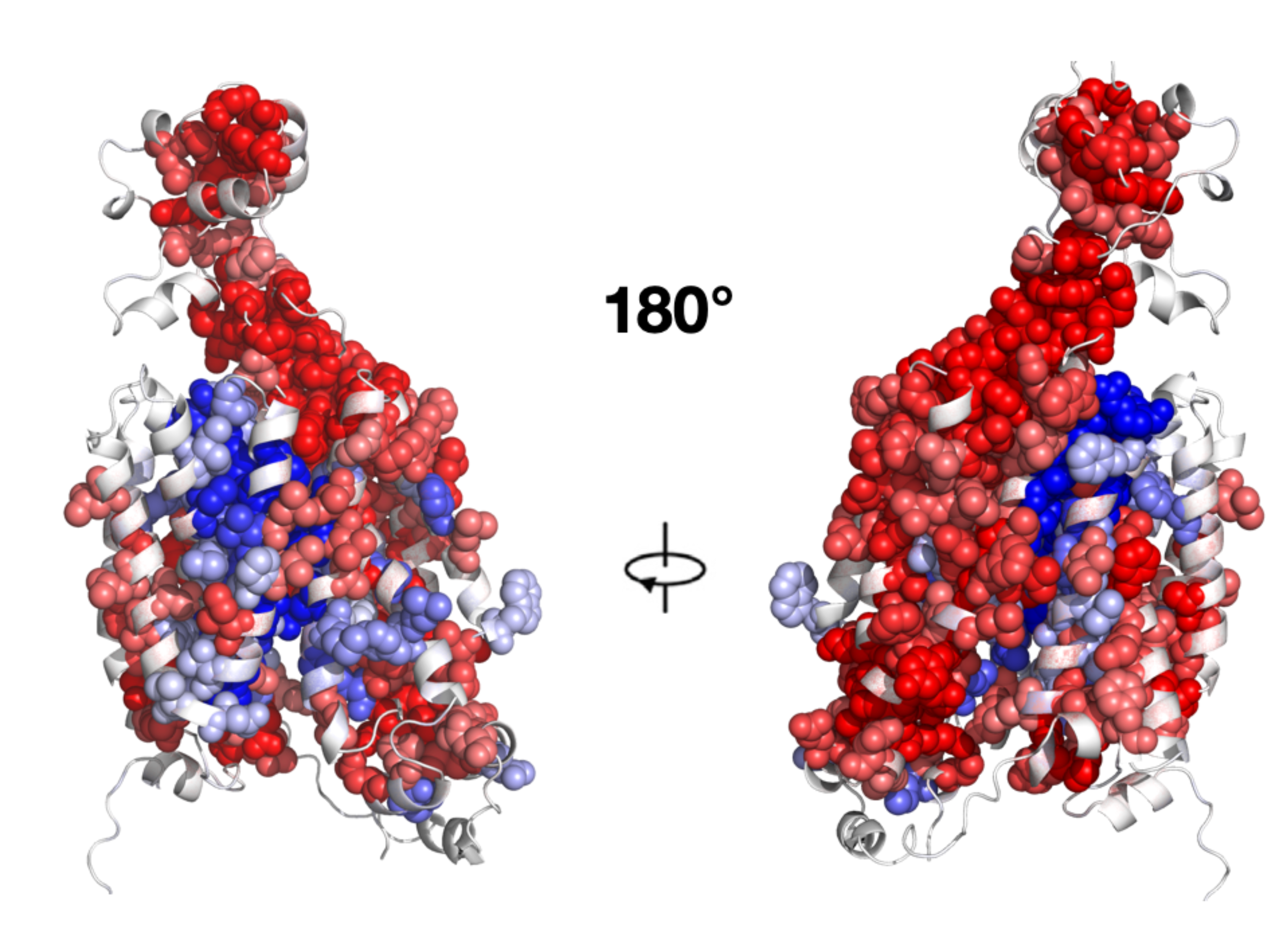
Our research
Membrane proteins are the main conduit through which cells interact with and sense the outside world. They determine key mechanisms of normal and abnormal physiology, interact with most drugs, and are involved in many diseases. Mutations in membrane proteins cause disease, disrupt treatments, and enable evolution. However, because membrane proteins are technically challenging to study, we understand far less about them than their cytosolic counterparts. We are pushing forward membrane protein biology by developing integrated models of how mutations, genes, and drugs change membrane protein structure, function, cell biology, and evolution.
Our long term goal is to develop a mechanistic molecular, cellular, and physiological understanding for how membrane proteins allows us to interact with the world around us, how they break to cause the diseases that ail us, and how we can better treat these diseases with therapeutics.
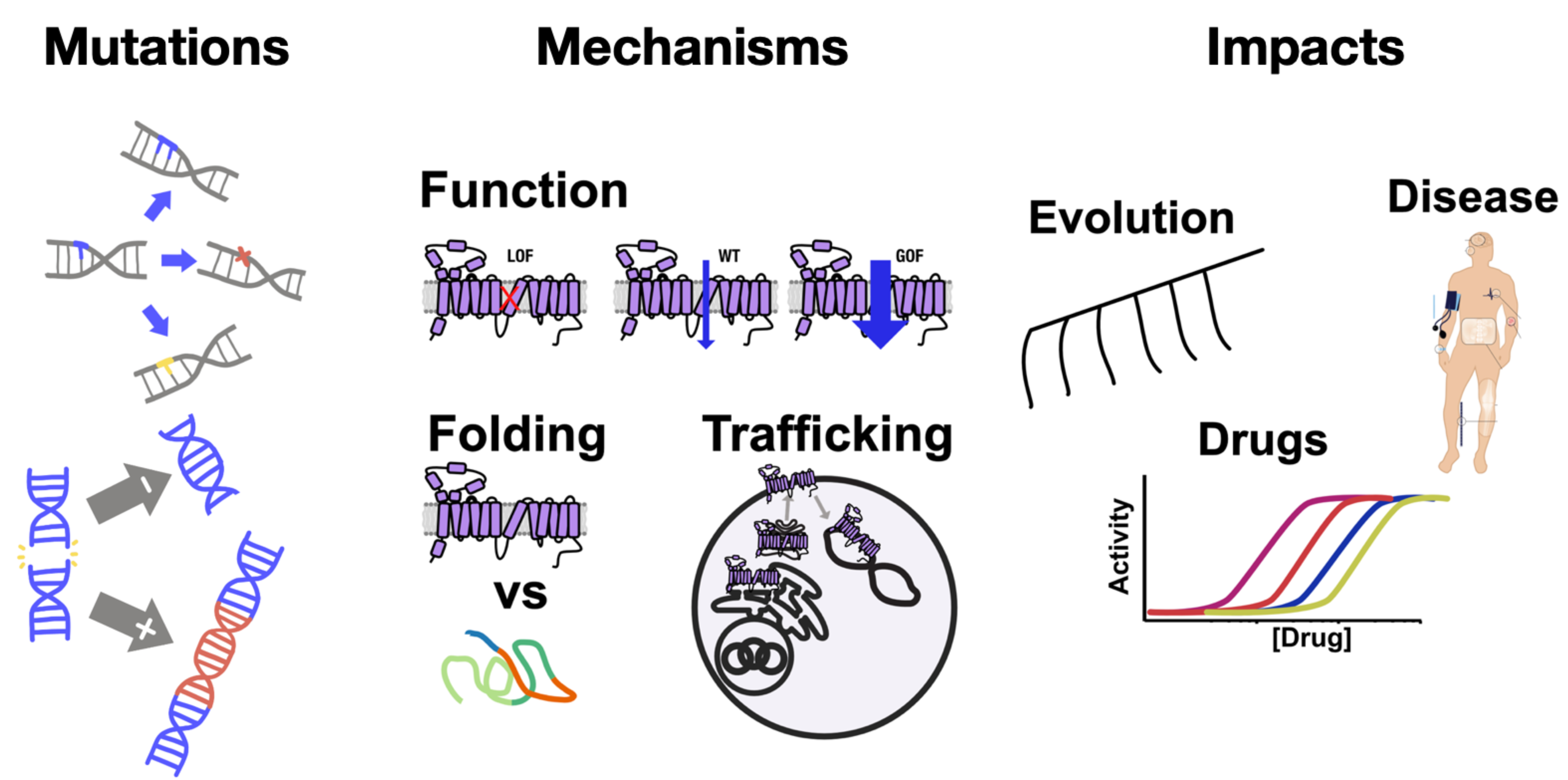
Develop foundational technologies for mechanistic genetic screening on membrane proteins
To understand how membrane protein residues change in evolution and disease, we screen libraries of mutations within membrane proteins. Currently, the mutational scanning community mostly focuses on single missense mutations leaving out other types of variation significant for disease and evolution, such as expression tuning synonymous mutations, architecture rearranging protein recombination, and combinations of multiple mutations. We will develop new library generation pipelines to systematically explore how synonymous variation, splice variants, protein recombination, and multiple mutations combine in membrane proteins to drive evolution, cause disease, and change pharmacology. We will use genetic variation and CRISPRi libraries to identify amino acids, motifs, and interacting genes that alter membrane protein’s function. To garner mechanistic insight directly from genetic screens, we will learn how perturbations impact membrane proteins. For this reason, we will develop a toolkit of membrane protein assays for measuring how perturbations impact folding, trafficking to organelles, interactions with partner proteins, downstream signaling, and overall functional and conformational states. We will integrate the results from these experiments using computational biology to develop biophysical models for how ligands, genes, and mutations change membrane protein folding, trafficking, function, and structure.
Molecular understanding for how receptors allow our cells to respond to sensation and therapeutics
GPCRs are cellular sensors and common drug targets. Through diverse mechanisms, mutations within GPCRs allow cells to evolve responses to new stimuli, break responses in disease, and disrupt drug efficacy. Complex mixtures of ligands activate GPCRs through multiple downstream effectors and signaling pathways making integrated biology necessary. Computational biologists develop models of GPCR activation and pharmacogenomics, however, we lack experimental approaches to test these models. Experimentalists typically focus on one GPCR at a time, with a couple of mutations and ligands, while looking at a small fraction of their impacts using conventional structural biology or cell biology methods. We are developing assays to identify how mutations, knocked-down genes, and ligands change GPCRs activation, signaling, and trafficking across all GPCRs. We are using these methods to learn how complex mixtures of ligands activate GPCRs, how sensory GPCRs change through evolution, and how mutations alter GPCR pharmacology and cause disease.
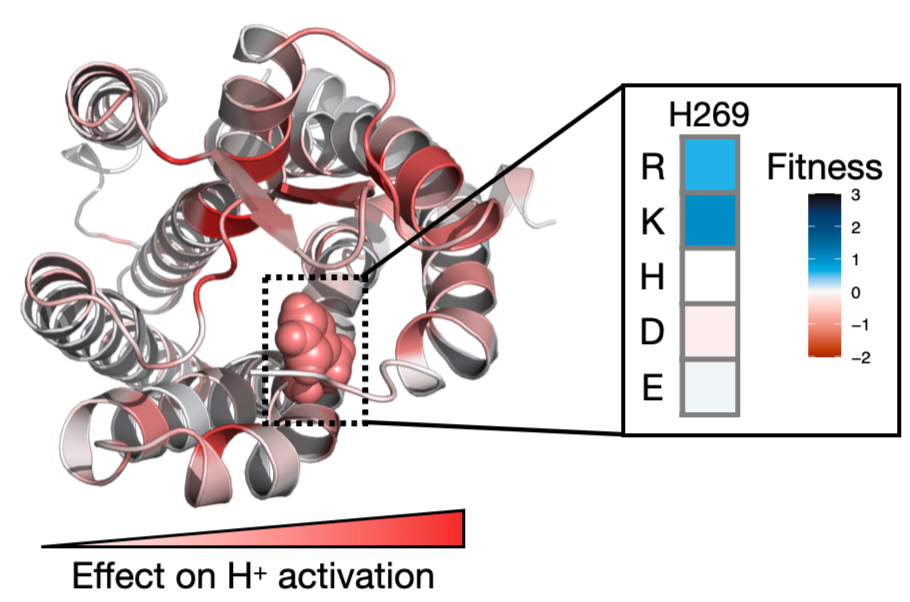
Discover the molecular mechanisms that underlie transporter substrate specificity
A large yet finite number of transporters evolved to move nearly infinite possible polar nutrients, small molecules, and drugs in and out of cells. Their role in exchanging solutes across the membrane makes transporters critical for physiology and pharmacology, illustrated by many disease-causing and therapeutic disrupting variants. Despite their importance, we do not know many transporters’ roles in physiology, what they transport, how they transport, or what determines a transporter’s substrate specificity. A lack of scalable mechanistic assays limits basic and applied transporter biology. We are addressing this challenge using genetic screens across the transporter-ome to uncover their roles in physiology. In follow-up experiments on specific transporters, we will do mechanistic mutational scans and structural experiments to develop structure-functional models of transporters. To determine pathogenicity and mechanisms of action, we will explore the functional impacts of human polymorphisms. From mechanistic genetic screens of membrane proteins, we are learning the physiological roles and structure-function mechanisms of an understudied class of proteins and actionable guidelines for clinical geneticists and pharmacists.